(Back to top)
Using snoTHAW
snoTHAW is a tool to help visualize snoRNA expression data from snoDB.
(Back to top)
Input
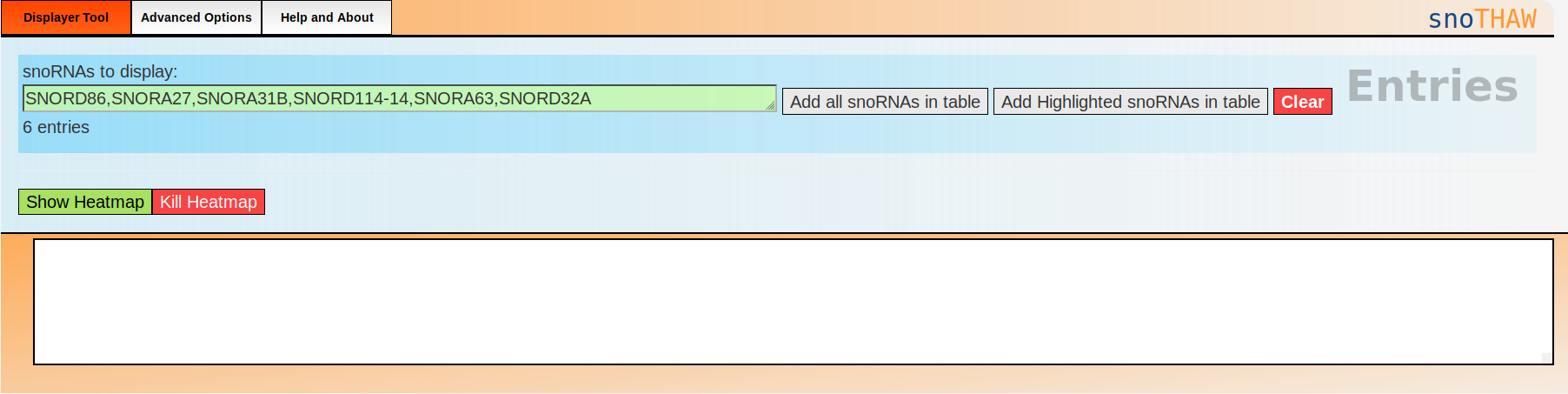
When you open snoTHAW, you should see the following:
-
Data Field:
The list of snoRNAs snoTHAW will be displaying once you press
"Show Heatmap"
(Back to top)
Output
If you press "Show Heatmap" with the example input selected, it will produce the following heatmap:

(Back to
top)
Adding data to the selection
Next to the Data Field you can see these 3 buttons:

-
Add all snoRNAs in table: Adds all the currently filtered entries in the table
-
Add Highlighted snoRNAs in table: By clicking a row in the table, it becomes
highlighted in blue
like
so:

You can use the Shift and Ctrl keys, just like you would for files in a
file
explorer, to make multiple and easy selections in the table.
-
Clear: Removes all entries from the Data Field.
You do not need to worry about inserting the same row more than once. snoTHAW will ensure that there are
no duplicates in the Data Field.
(Back to top)
Heatmap Production
At the bottom of the tool section, you will see the following buttons:

-
Show Heatmap:
Will produce the heatmap visualization.
You can press the same button
at the bottom of this section
to produce the example heatmap.
-
Kill Heatmap:
Will erase the heatmap object to save memory.
(Back to top)
Interacting with the Heatmap
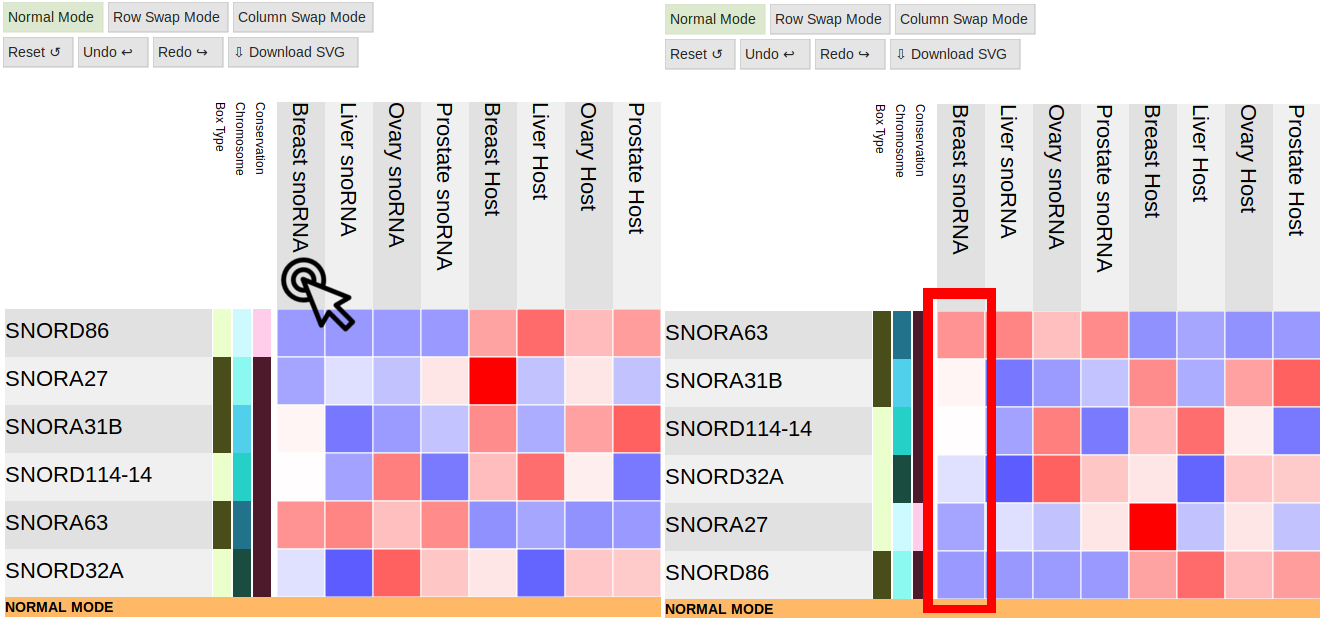
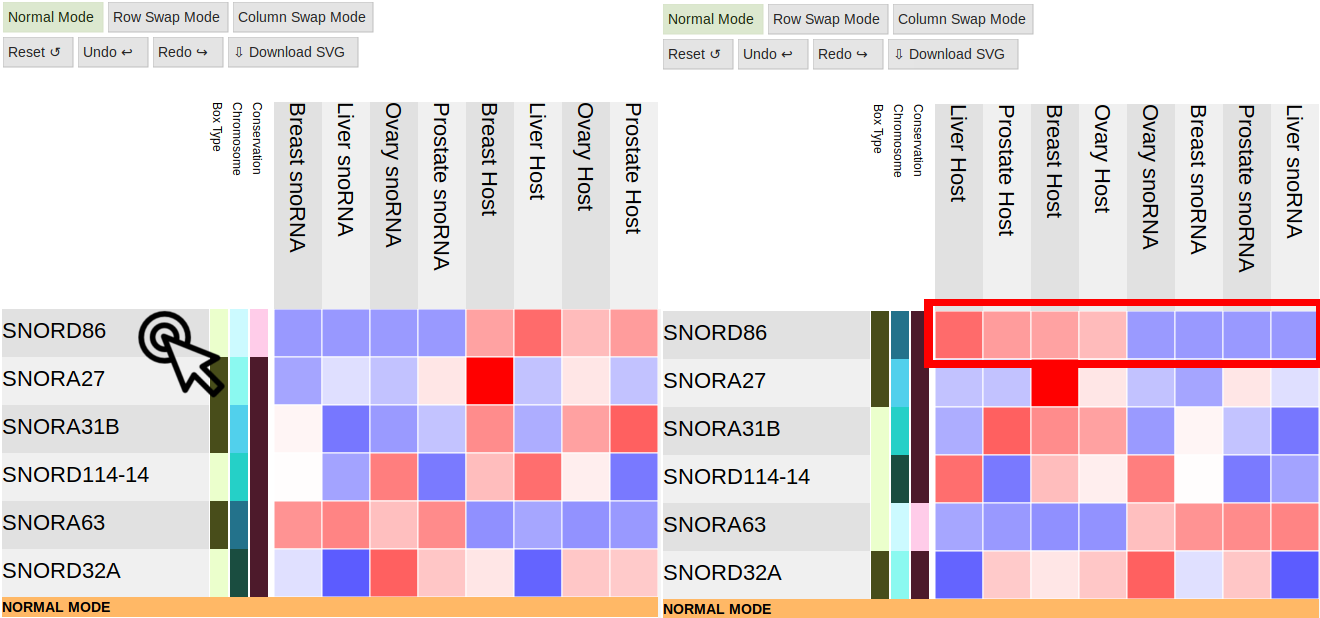
Double-Clicking on columns and rows
Double-Clicking on a column or row label will reorganize the heatmap to show that row or column's data in
decreasing order.
-
Columns
-
Rows
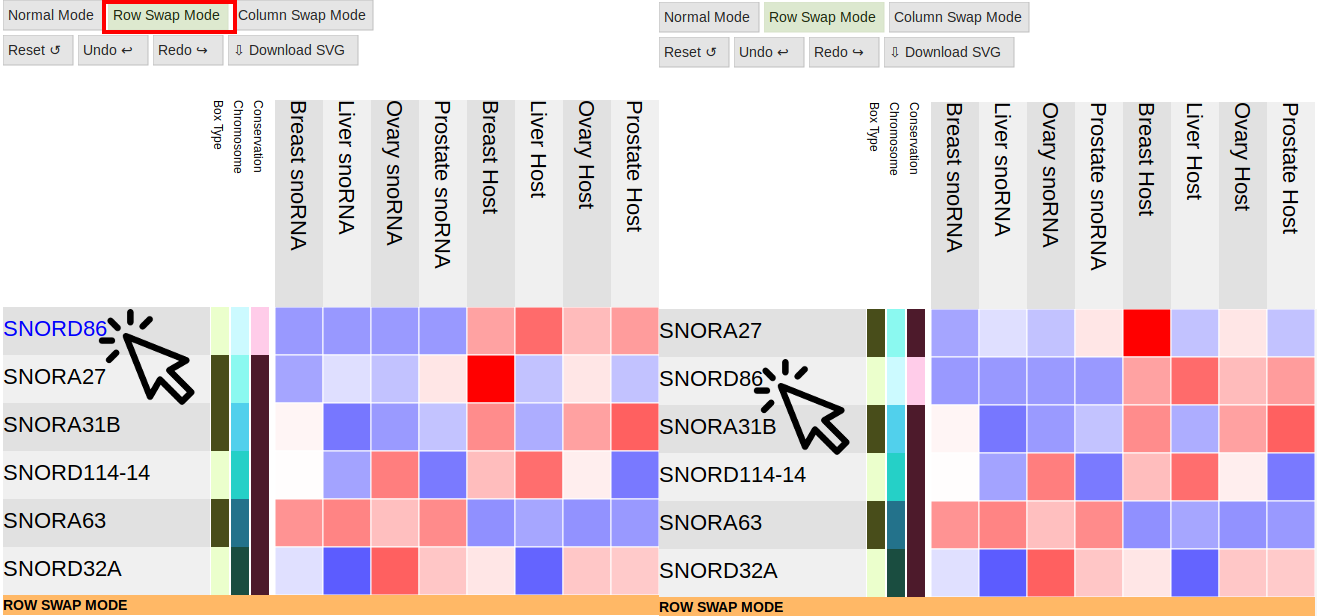
Row Swap Mode and Column Swap Mode
Pressing these buttons will allow you to swap two rows or columns to reorganize the table however you want:
-
Columns

-
Rows
Double-Clicking on a Characteristics Square
Doing so will group the characteristics together and make the group you clicked on appear first: